阅读并同意隐私协议、MTA和使用条款后进行下一步!
CoronaViruses Sequence Comparison


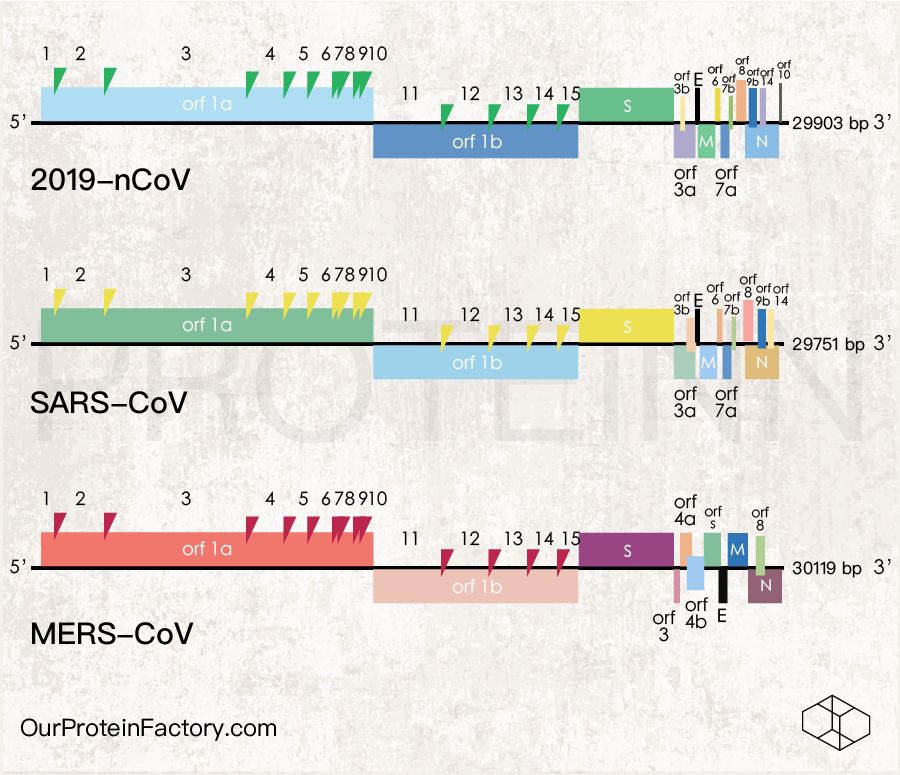
Figure 1.Schematics of 2019-nCoV, SARS-CoV, MERS-CoV genome, ORF and protein.
The following analysis is based on the most up-to-date 2019-nCoV genome sequence Refseq: NC_045512 [1].
Genome
For genome sequence, the 2019-nCoV RNA (29903 bases) shares 80.08% homology to SARS-CoV (29751 bases, Refseq. NC_004718), 58.79% to MERS-CoV (30119 bases, Refseq. NC_019843). The homology between SARS-CoV and MERS-CoV genome sequences is 47.07%.
Table 1.Orthogonal Array of 3 Genomic RNAs Homology.
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
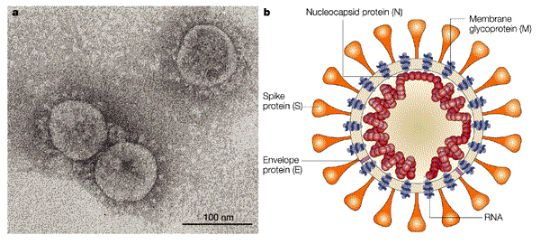
Table 2. Number of ORF and Protein in the 3 coronaviruses.
|
Number |
ORF |
Protein |
|
2019-nCoV |
15 |
28 |
|
SARS-nCoV |
14 |
27 |
|
MERS-nCoV |
11 |
24 |
Table 3. Proteins encoded in the 3 coronaviruses.
|
Protein\Residue Number |
2019-nCoV |
SARS-nCoV |
MERS-nCoV |
|
5' UTR |
/ |
/ |
/ |
|
Host translation inhibitor NSP1 |
180 |
180 |
193 |
|
Non-structural protein 2 |
638 |
638 |
660 |
|
Papain-like proteinase |
1945 |
1922 |
1887 |
|
Non-structural protein 4 |
500 |
500 |
507 |
|
3C-like proteinase |
306 |
306 |
306 |
|
Non-structural protein 6 |
290 |
290 |
292 |
|
Non-structural protein 7 |
83 |
83 |
83 |
|
Non-structural protein 8 |
198 |
198 |
199 |
|
Non-structural protein 9 |
113 |
113 |
110 |
|
Non-structural protein 10 |
139 |
139 |
140 |
|
RNA-directed RNA polymerase |
932 |
932 |
933 |
|
Helicase |
601 |
601 |
598 |
|
Guanine-N7 methyltransferase |
527 |
527 |
524 |
|
Uridylate-specific endoribonuclease |
346 |
346 |
343 |
|
2'-O-methyltransferase |
298 |
210 |
303 |
|
Spike glycoprotein (S) |
1273 |
1255 |
1353 |
|
orf 3a |
275 |
274 |
103 |
|
Envelope protein (E) |
75 |
76 |
82 |
|
Membrane glycoprotein (M) |
222 |
221 |
219 |
|
orf 4a |
/ |
/ |
109 |
|
orf 4b |
/ |
/ |
246 |
|
orf 5 |
/ |
/ |
224 |
|
orf 6 |
61 |
63 |
/ |
|
orf 7a |
121 |
122 |
/ |
|
orf 8 |
121 |
84 |
112 |
|
Nucleocapsid phosphoprotein (N) |
419 |
422 |
413 |
|
orf9b |
/ |
98 |
/ |
|
orf 10 |
38 |
/ |
/ |
|
3' UTR |
/ |
/ |
/ |
a. The length of each protein is displayed. .
b. “/” means that no protein is encoded.
Table 4. Protein Homology in the 3 coronaviruses.
|
Protein Name\Identity |
2019-n/SARS |
2019-n/MERS |
SARS/MERS |
|
Host translation inhibitor NSP1 |
84.4% |
25.7% |
25.0% |
|
Non-structural protein 2 |
68.3% |
21.4% |
21.9% |
|
Papain-like proteinase |
76.7% |
33.2% |
32.9% |
|
Non-structural protein 4 |
80.0% |
39.6% |
38.2% |
|
3C-like proteinase |
96.8% |
50.8% |
51.8% |
|
Non-structural protein 6 |
87.2% |
34.8% |
34.8% |
|
Non-structural protein 7 |
98.8% |
55.4% |
55.4% |
|
Non-structural protein 8 |
97.5% |
52.8% |
52.3% |
|
Non-structural protein 9 |
97.4% |
53.6% |
53.6% |
|
Non-structural protein 10 |
97.1% |
59.0% |
59.0% |
|
RNA-directed RNA polymerase |
96.4% |
72.6% |
72.3% |
|
Helicase |
99.8% |
71.6% |
71.6% |
|
Guanine-N7 methyltransferase |
95.1% |
63.4% |
63.2% |
|
Uridylate-specific endoribonuclease |
88.7% |
51.8% |
50.0% |
|
2'-O-methyltransferase |
93.3% |
66.1% |
65.1% |
|
Spike glycoprotein (S) |
77.7% |
33.3% |
33.7% |
|
orf 3a |
72.9% |
7.2% |
8.3% |
|
orf 3b |
7.8% |
/ |
/ |
|
Envelope protein (E) |
93.2% |
36.5% |
36.8% |
|
Membrane glycoprotein (M) |
91.0% |
40.2% |
42.0% |
|
orf 7a |
86.0% |
/ |
/ |
|
orf 7b |
83.3% |
/ |
/ |
|
orf 8 |
57.0% |
17.1% |
16.2% |
|
Nucleocapsid phosphoprotein (N) |
91.2% |
49.9% |
49.6% |
|
orf 9a |
73.2% |
/ |
/ |
|
orf 14 |
77.1% |
/ |
/ |
References.
https://www.ncbi.nlm.nih.gov/nuccore/1798174254