阅读并同意隐私协议、MTA和使用条款后进行下一步!


• Introduction
• Specimens
• Reagents, Supplies and Equipment Requirements
• Nucleic Acid Extraction
• Quality Control
• rRT-PCR Assays
• Interpreting Test Results
• Assay Limitations
• Contact Information
Purpose:This document describes the use of real-time RT PCR (rRT-PCR) assays for the in vitro qualitative detection of 2019-Novel Coronavirus (2019-nCoV) in respiratory specimens and sera. The 2019-nCoV primer and probe sets are designed for the universal detection of SARS-like coronaviruses (N3 assay) and for specific detection of 2019-nCoV (N1 and N2 assays).
Protocol Use Limitations: The rRT-PCR assays described here have not been validated for platforms or chemistries other than those described in this document.
Wear appropriate personal protective equipment (e.g. gowns, gloves, eye protection) when working with clinical specimens. Specimen processing should be performed in a certified class II biological safety cabinet following biosafety level 2 or higher guidelines.
For more information, refer to:
• Interim Guidelines for Collecting, Handling, and Testing Clinical Specimens from Persons Under Investigation (PUIs) for 2019 Novel Coronavirus (2019-nCoV)
https://www.cdc.gov/coronavirus/2019-nCoV/lab/guidelines-clinical-specimens.html
• Biosafety in Microbiological and Biomedical Laboratories 5th edition available athttp://www.cdc.gov/biosafety/publications/
• Respiratory specimens including: nasopharyngeal or oropharyngeal aspirates or washes, nasopharyngeal or oropharyngeal swabs, broncheoalveolar lavage, tracheal aspirates, and sputum.
• Swab specimens should be collected only on swabs with a synthetic tip (such as polyester or Dacron®) with aluminum or plastic shafts. Swabs with calcium alginate or cotton tips with wooden shafts are not acceptable.
• Serum
• Specimens can be stored at 4℃ for up to 72 hours after collection.
• If a delay in extraction is expected, store specimens at -70℃ or lower.
• Extracted nucleic acids should be stored at -70℃ or lower.
• Specimens not kept at 2-4℃ (≤4 days) or frozen at -70℃ or below.
• Incomplete specimen labeling or documentation.
• Inappropriate specimen type.
• Insufficient specimen volume.
Disclaimer: Names of vendors or manufacturers are provided as examples of suitable product sources. Inclusion does not imply endorsement by the Centers for Disease Control and Prevention.
• rRT-PCR primer/probe sets
• Positive template control
• TaqPath™ 1-Step RT-qPCR Master Mix, CG (ThermoFisher; cat # A15299 or A15300)
• Molecular grade water, nuclease-free
• Disposable powder-free gloves
• P2/P10, P200, and P1000 aerosol barrier tips
• Sterile, nuclease-free 1.5 mL microcentrifuge tubes
• 0.2 mL PCR reaction tube strips or 96-well real-time PCR reaction plates and optical 8-cap strips
• Laboratory marking pen
• Cooler racks for 1.5 microcentrifuge tubes and 96-well 0.2 mL PCR reaction tubes
• Racks for 1.5 ml microcentrifuge tubes
• Acceptable surface decontaminants
◦ DNAZapTM (Life Technologies, cat. #AM9890)
◦ DNA AwayTM (Fisher Scientific; cat. #21-236-28)
◦ RNAse AwayTM (Fisher Scientific; cat. #21-236-21
◦ 10% bleach (1:10 dilution of commercial 5.25-6.0% sodium hypochlorite)
• PCR Work Station [UV lamp; Laminar flow (Class 100 HEPA filtered)]
• Vortex mixer
• Microcentrifuge
• Micropipettes (2 or 10 µl, 200 µl and 1000 µl)
• Multichannel micropipettes (5-50 µl)
• 2 x 96-well cold blocks
• -20℃ (nonfrost-free) and -70℃ freezers; 4℃ refrigerator
• Real-time PCR detection system
• Nucleic acid extraction system
• Performance of rRT-PCR amplification based assays depends on the amount and quality of sample template RNA. RNA extraction procedures should be qualified and validated for recovery and purity before testing specimens.
• Commercially available extraction procedures that have been shown to generate highly purified RNA when following manufacturer’s recommended procedures for sample extraction include: bioMérieux NucliSens® systems, QIAamp® Viral RNA Mini Kit, QIAamp® MinElute Virus Spin Kit or RNeasy® Mini Kit (QIAGEN), EZ1 DSP Virus Kit (QIAGEN), Roche MagNA Pure Compact RNA Isolation Kit, Roche MagNA Pure Compact Nucleic Acid Isolation Kit, and Roche MagNA Pure 96 DNA and Viral NA Small Volume Kit, and Invitrogen ChargeSwitch® Total RNA Cell Kit.
• Retain residual specimen and nucleic extract and store immediately at -70℃.
• Only thaw the number of specimen extracts that will be tested in a single day. Do not freeze/thaw extracts more than once before testing.
Due to the sensitivity of rRT-PCR, these assays should be conducted using strict quality control and quality assurance procedures. Following these guidelines will help minimize chance of false-positive amplification.
• Personnel must be familiar with the protocol and instruments used.
• Maintain separate areas and dedicated equipment (e.g., pipettes, microcentrifuges) and supplies (e.g., microcentrifuge tubes, pipette tips, gowns and gloves) for assay reagent setup and handling of extracted nucleic acids.
• Work flow must always be from the clean area to the dirty area.
• Wear clean disposable gowns and new, previously unworn, powder-free gloves during assay reagent setup and handling of extracted nucleic acids. Change gloves whenever contamination is suspected.
• Store primer/probes and enzyme master mix at appropriate temperatures (see package inserts). Do not use reagents beyond their expiry dates.
• Keep reagent tubes and reactions capped as much as possible.
• Clean and decontaminate surfaces.
• Do not bring extracted nucleic acid or PCR products into the assay setup area.
• Use aerosol barrier (filter) pipette tips only.
• Use PCR plate strip caps only. Do not use PCR plate sealing film.
• Assay controls should be run concurrently with all test samples.
• PTC – positive template control with an expected Ct value range
• NTC – negative template control added during rRT-PCR reaction set-up
• HSC – human specimen extraction control extracted concurrently with the test samples; provides a nucleic acid extraction procedural control and a secondary negative control that validates the nucleic extraction procedure and reagent integrity
• RP – all clinical samples should be tested for human RNAse P (RNP) gene to assess specimen quality
• Note: Keep running logs of PTC performance. After each rRT-PCR run of clinical samples, the control Ct values should be recorded.
rRT-PCR Primers/Probe Sets (see package insert if provided by CDC)
• Precautions: These reagents should only be handled in a clean area and stored at appropriate temperatures (see below) in the dark. Freeze-thaw cycles should be avoided. Maintain cold when thawed.
• Using aseptic technique, suspend dried reagents in 1.5 mL nuclease-free water and allow to rehydrate for 15 min at room temperature in the dark.
• Mix gently and aliquot primers/probe in 300 μL volumes into 5 pre-labeled tubes. Store a single aliquot of primers/probe at 2-8oC in the dark. Do not refreeze (stable for up to 4 months). Store remaining aliquots at ≤-20oC in a non-frost-free freezer.
• Positive Template Control (PTC) (if using CDC positive control, nCoV PC, see package insert provided by CDC)
• Precautions: This reagent should be handled with caution in a dedicated nucleic acid handling area to prevent possible contamination. Freeze-thaw cycles should be avoided. Maintain on ice when thawed.
• Used to assess performance of rRT-PCR assays. Resuspend dried reagent in each tube in 1 mL of nuclease-free water to achieve the proper concentration. Make single use aliquots (approximately 30 μL) and store at ≤ -70℃.
• Thaw a single aliquot of diluted positive control for each experiment and hold on ice until adding to plate. Discard any unused portion of the aliquot.
• The nCoVPC also contains human DNA that serves as the positive control for the RP assay.
1、Clean and decontaminate all work surfaces, pipets, centrifuges and other equipment prior to use using RNase Away® or 10% freshly prepared bleach.
2、Turn on AB 7500 Fast DX and allow block to reach optimal temperature.
3、Perform plate set up and select cycling protocol on the instrument.
4、Instrument Settings: Detector (FAM); Quencher (None); Passive Reference: (None); Run Mode: (Standard); Sample Volume (20 µL)
Equipment preparation
* Fluorescence data (FAM) should be collected during the 55℃ incubation step.
Note: Plate set-up configuration can vary with the number of specimens and work day organization. NTCs and nCoVPCs must be included in each run.
1、In the reagent set-up room clean hood, place r4X Master Mix and primer/probes on ice or cold-block. Keep cold during preparation and use.
2、Thaw 4X Reaction Mix prior to use.
3、Mix 4X Master Mix and primer/probes by inversion 5 times.
4、Briefly centrifuge 4X Master Mix and primers/probes and return to cold block.
5、Label one 1.5 mL microcentrifuge tube for each primer/probe set.
6、Determine the number of reactions (N) to set up per assay. It is necessary to make excess reaction mix for the NTC, nCoVPC, and RP reactions and for pipetting error. Use the following guide to determine N:
If number of samples (n) including controls equals 1 through 14,
then N = n + 1
If number of samples (n) including controls is 15 or greater,
then N = n + 2
7、For each primer/probe set, calculate the amount of each reagent to be added for each reaction mixture (N = # of reactions).
TaqPath™ 1-Step RT-qPCR Master Mix
8、Dispense reagents into each respective labeled 1.5 mL microcentrifuge tube. After addition of the reagents, mix reaction mixtures by pipetting up and down. Do not vortex.
9、Centrifuge for 5 seconds to collect contents at the bottom of the tube, and then place the tube in a cold rack.
10、Set up reaction strip tubes or plates in a 96-well cooler rack.
11、Dispense 15 µL of each master mix into the appropriate wells going across the row as shown below (Figure 1):
Figure 1: Example of Reaction Master Mix Plate Set-Up
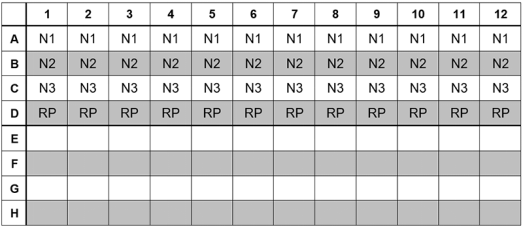
12、Prior to moving to the nucleic acid handling area, prepare the No Template Control (NTC) reactions for column #1 in the assay preparation area.
13、Pipette 5 µL of nuclease-free water into the NTC sample wells. Securely cap NTC wells before proceeding.
14、Cover the entire reaction plate and move the reaction plate to the specimen nucleic acid handling area.
1、Gently vortex nucleic acid sample tubes for approximately 5 seconds.
2、After centrifugation, place extracted nucleic acid sample tubes in the cold rack.
3、Samples should be added to column 2-11 (column 1 and 12 are for controls) to the specific assay that is being tested as illustrated in Figure 2. Carefully pipette 5.0 µL of the first sample into all the wells labeled for that sample (i.e. Sample “S1” down column #2). Keep other sample wells covered during addition. Change tips after each addition.
4、Securely cap the column to which the sample has been added to prevent cross contamination and to ensure sample tracking.
5、Change gloves often and when necessary to avoid contamination.
6、Repeat steps #3 and #4 for the remaining samples.
7、If necessary, add 5 µL of Human Specimen Control (HSC) extracted sample to the HSC wells (Figure 2, column 11). Securely cap wells after addition
8、Cover the entire reaction plate and move the reaction plate to the positive template control handling area.
9、Pipette 5 µL of nCoVPC RNA to the sample wells of column 12 (Figure 2). Securely cap wells after addition of the control RNA.
10、NOTE:If using 8-tube strips, label the TAB of each strip to indicate sample position. DO NOT LABEL THE TOPS OF THE REACTION TUBES!
11、Briefly centrifuge reaction tube strips for 10-15 seconds. After centrifugation return to cold rack.
12、NOTE: If using 96-well plates, centrifuge plates for 30 seconds at 500 x g, 4℃.
Figure 2. 2019-nCoV rRT-PCR Diagnotic Panel:Example of Sample and Control Set-up
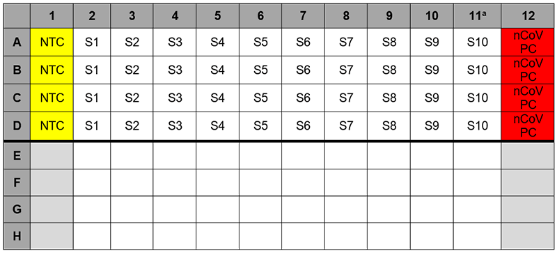
aReplace the sample in this column with extracted HSC if necessary
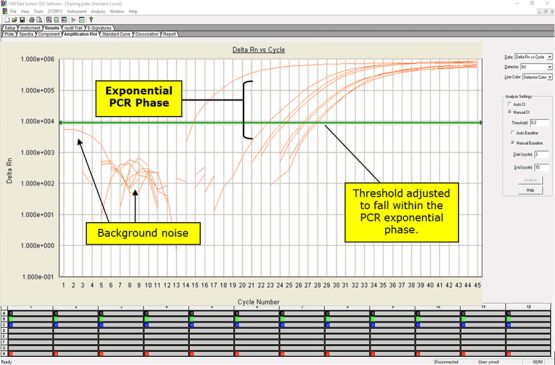
• After completion of the run, save and analyze the data following the instrument manufacturer’s instructions. Analyses should be performed separately for each target using a manual threshold setting. Thresholds should be adjusted to fall within exponential phase of the fluorescence curves and above any background signal (refer to Figure 3). The procedure chosen for setting the threshold should be used consistently.
Figure 3. Amplification Plot Window
• NTCs should be negative and not exhibit fluorescence growth curves that cross the threshold line.
• If a false positive occurs with one or more of the primer and probe NTC reactions, sample contamination may have occurred.
• Invalidate the run and repeat the assay with stricter adherence to the procedure guidelines.
• PTC reaction should produce a positive result with an expected Ct value for each target included in the test.
• If expected positive reactivity is not achieved, invalidate the run and repeat the assay with stricter adherence to procedure guidelines.
• Determine the cause of failed PTC reactivity, implement corrective actions, and document results of the investigation and corrective actions.
• Do not use PTC reagents that do not generate expected result.
• RP should be positive at or before 35 cycles for all clinical samples and HSC, thus indicating the presence of sufficient nucleic acid from human RNase P gene and that the specimen is of acceptable quality.
• Failure to detect RNase P in HSC may indicate:
• Improper assay set up and execution
• Reagent or equipment malfunction
• Detection of RNase P in HSC but failure to detect RNase P in any of the clinical samples may indicate:
• Improper extraction of nucleic acid from clinical materials resulting in loss of nucleic acid or carry-over of PCR inhibitors from clinical specimens
• Absence of sufficient human cellular material in sample to enable detection
• HSC should be negative for 2019-nCoV specific primer/probe sets.
• If any 2019-nCoV specific primer/probes exhibit a growth curve that crosses the threshold line, interpret as follows:
• Contamination of nucleic acid extraction reagents may have occurred. Invalidate the run and confirm reagent integrity of nucleic acid extraction reagents prior to further testing.
• Cross contamination of samples occurred during nucleic acid extraction procedures or assay setup. Invalidate the run and repeat the assay with stricter adherence to procedure guidelines.
• When all controls exhibit the expected performance, a specimen is considered negative if all 2019-nCoV markers (N1, N2, N3) cycle threshold growth curves DO NOT cross the threshold AND the RNase P growth curve DOES cross the threshold line.
• When all controls exhibit the expected performance, a specimen is considered positive for 2019-nCoV if all markers (N1, N2, N3) cycle threshold growth curve crosses the threshold line. The RNase P may or may not be positive as described above, but the 2019-nCoV result is still valid.
• When all controls exhibit the expected performance and the growth curves for the 2019-nCoV markers (N1, N2, N3) AND the RNase P marker DO NOT cross the cycle threshold growth curve, the result is invalid. The extracted RNA from the specimen should be re-tested. If residual RNA is not available, re-extract RNA from residual specimen and re-test. If the re-tested sample is negative for all markers and all controls exhibit the expected performance, the result is “Invalid.”
• When all controls exhibit the expected performance and the cycle threshold growth curve for any one or two markers, (N1, N2, N3) but not all three crosses the threshold line the result is inconclusive for 2019-nCoV. Re-extract RNA from residual specimen and re-test.
2019-nCoV rRT-PCR Diagnostic Panel Results Interpretation
• Analysts should be trained and familiar with testing procedures and interpretation of results prior to performing the assay.
• A false negative result may occur if inadequate numbers of organisms are present in the specimen due to improper collection, transport or handling.
• RNA viruses in particular show substantial genetic variability. Although efforts were made to design rRT-PCR assays to conserved regions of the viral genomes, variability resulting in mis-matches between the primers and probes and the target sequences can result in diminished assay performance and possible false negative results.
Suggestions and questions concerning this procedure may be sent to:
respvirus@cdc.govPage last reviewed: February 5, 2020
Content source: National Center for Immunization and Respiratory Diseases (NCIRD), Division of Viral Diseases