阅读并同意隐私协议、MTA和使用条款后进行下一步!


Supported by the world-leading Cell-Free Protein Synthesis (CFPS) technology D2P, OurProteinFactory now offers Cell-Free expressed 25 proteins from the 2019-nCoV coronavirus. Expressed in ProteinFactory Standard package, His-Monster beads purified, His- and/or eGFP-tagged, spike proteins (S1, S2, S-RBD), N protein, M protein, E protein, 3C-Like Protease, RdRp etc. are all in stock.
基于世界领先的无细胞蛋白质合成(Cell-Free Protein Synthesis CFPS)技术 D2P 的支持下,OurProteinFactory 现提供来自2019-nCoV冠状样病毒的,通过无细胞表达制造的25种蛋白质。 以 ProteinFactory Standard 标准装表达,His-Monster beads 磁珠纯化,具有 His- 和/或 eGFP- 标签的,多种病毒蛋白如:刺突蛋白(S1,S2,S-RBD),N蛋白,M蛋白,E蛋白,3C-Like蛋白酶,RdRp等,现货供应。
Using pD2P expression vector, all 2019-nCoV proteins coding sequences are codon-optimized, gene synthesized, no homology to viral RNA, no cell involved, no cross-infectivity to live virus, are available to advance the global COVID-19 research and drug development.
所有2019-nCoV蛋白的DNA编码序列均经过密码子优化,基因合成,使用pD2P载体,完全体外表达,无细胞参与,与活病毒核酸序列无交叉,无感染性,助力于全球冠状样病毒疫情监测、防御和治疗的研发。
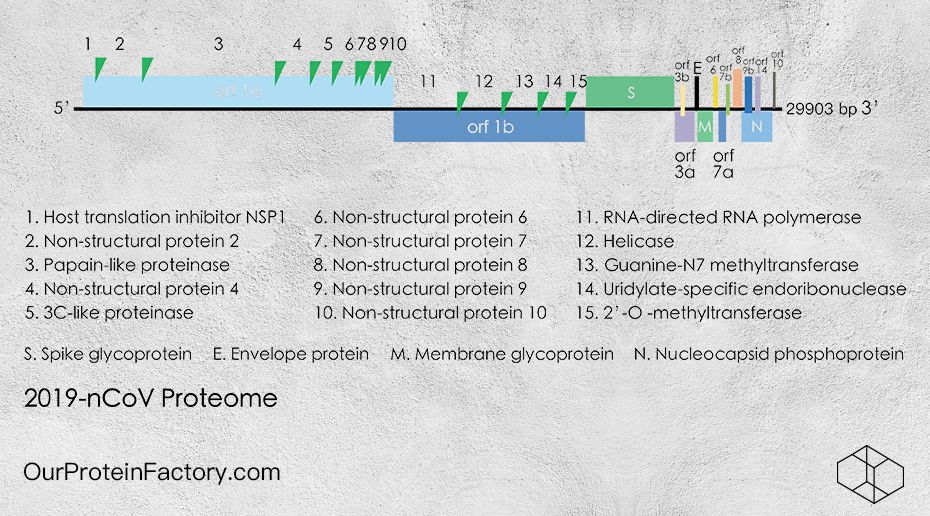
The 2019-nCoV RNA (29903 bases) encodes 15 Open Reading Frames (ORFs)。Among them, orf1ab is the longest (21330 bases). During protein translation, a codon frame-shift (at position 13468) results in producing two polyproteins as pp1a, pp1ab. [2] These polyproteins are cleaved into 15 proteins. The entire 2019-nCoV RNA encodes 28 proteins in total.
2019-nCoV冠状样病毒RNA(29903个碱基)共含有15个开放阅读框(ORFs)。orf1ab 长度最长(21330 个碱基)。orf1ab 翻译时通过一个中部密码子翻译框位移 (位于13468位),产生两个大的多体蛋白:pp1a和pp1ab。[2] 这些多体蛋白被分别剪切,形成共1 5种蛋白质。整个2019-nCoV病毒RNA共编码28种蛋白质。
The following protein analysis is based on the most up-to-date 2019-nCoV genome sequence Refseq: NC_045512. [1]
以下蛋白质分析基于NCBI数据库中最新更新的2019-nCoV基因组序列,Refseq: NC_045512. [1]
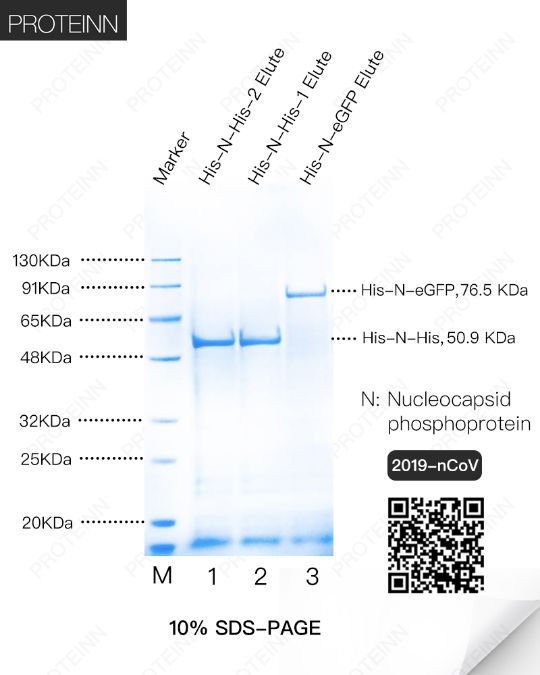
N: Nucleocapsid phosphoprotein
Function: Packages the positive strand viral genome RNA into a helical ribonucleocapsid (RNP) and plays a fundamental role during virion assembly through its interactions with the viral genome and membrane protein M. Plays an important role in enhancing the efficiency of subgenomic viral RNA transcription as well as viral replication.
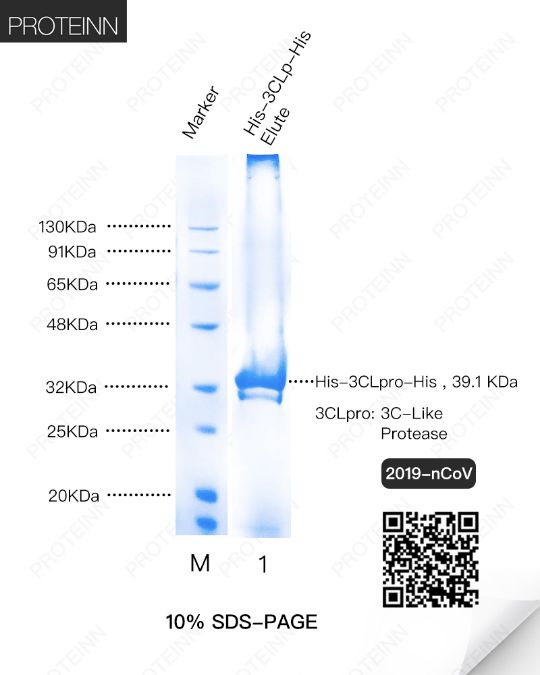
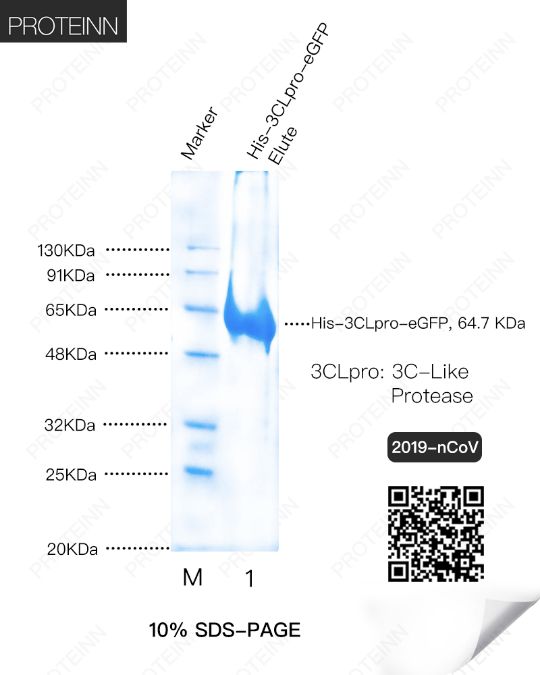
3CLp: 3C-Like Protease
Function: also named as Mpro (Main Protease), cleaves the C-terminus of replicase polyprotein at 11 sites. Recognizes substrates containing the core sequence [ILMVF]-Q-|-[SGACN]. Also able to bind an ADP-ribose-1''-phosphate (ADRP).
NendoU: Uridylate-specific endoribonuclease
Function: Mn2+-dependent, uridylate-specific enzyme, which leaves 2'-3'-cyclic phosphates 5' to the cleaved bond.
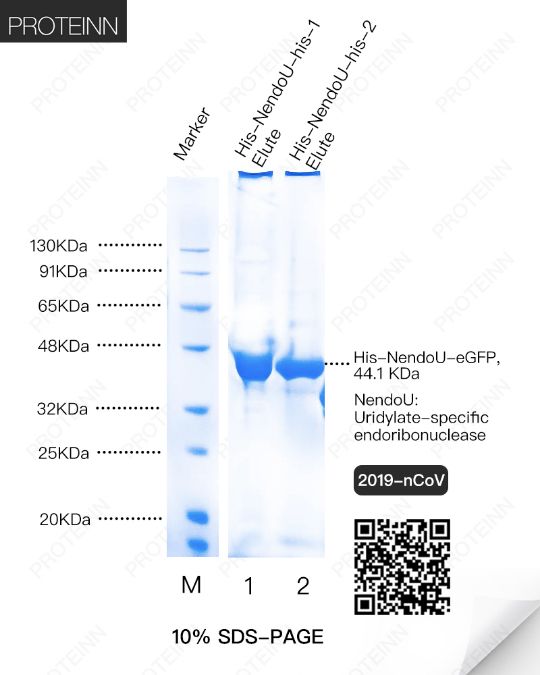
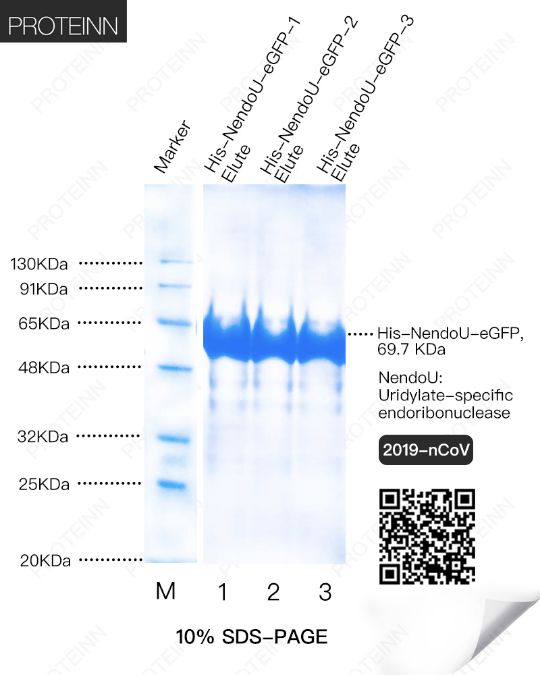
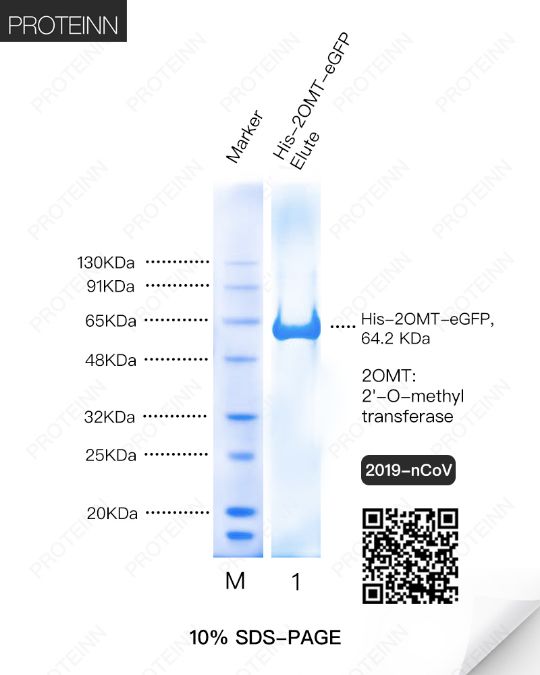
2OMT: 2'-O-methyltransferase
Function: Methyltransferase that mediates mRNA cap 2'-O-ribose methylation to the 5'-cap structure of viral mRNAs. N7-methyl guanosine cap is a prerequisite for binding of nsp16. Therefore plays an essential role in viral mRNAs cap methylation which is essential to evade immune system.
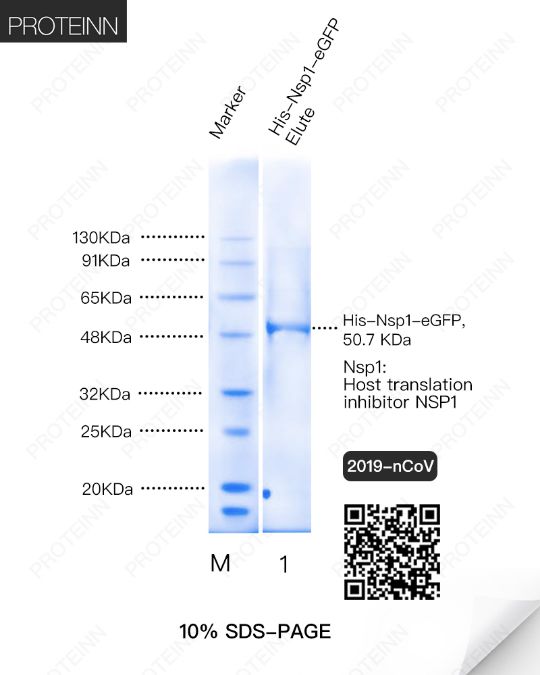
Nsp1: Host translation inhibitor NSP1
Function: Inhibits host translation by interacting with the 40S ribosomal subunit. The nsp1-40S ribosome complex further induces an endonucleolytic cleavage near the 5'UTR of host mRNAs, targeting them for degradation. Viral mRNAs are not susceptible to nsp1-mediated endonucleolytic RNA cleavage thanks to the presence of a 5'-end leader sequence and are therefore protected from degradation. By suppressing host gene expression, nsp1 facilitates efficient viral gene expression in infected cells and evasion from host immune response.
Nsp2: Non-Structural Protein 2
Function: May play a role in the modulation of host cell survival signaling pathway by interacting with host PHB and PHB2. Indeed, these two proteins play a role in maintaining the functional integrity of the mitochondria and protecting cells from various stresses.
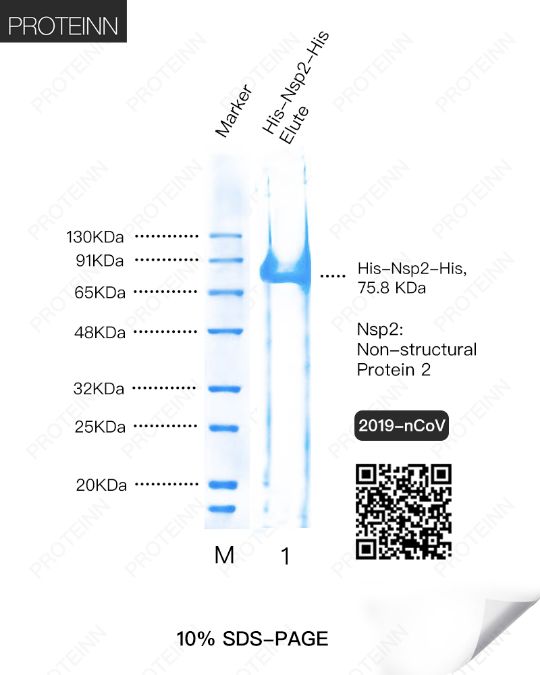
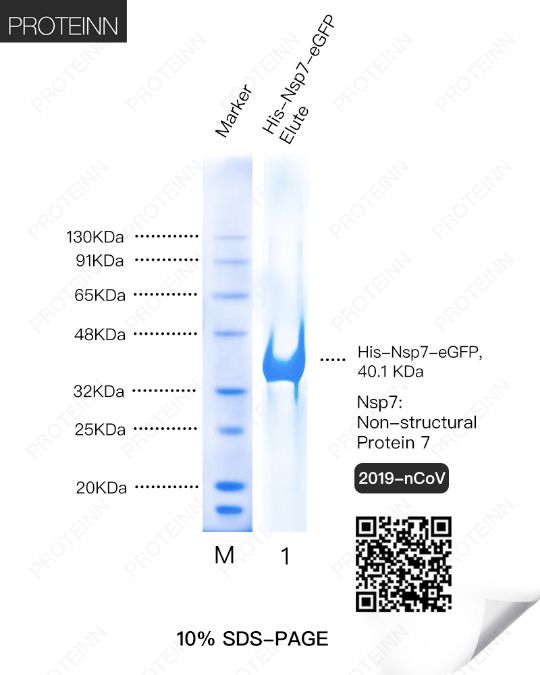
Nsp7: Non-Structural Protein 7
Function: acting as a primase. Alternatively, may synthesize substantially longer products than oligonucleotide primers.
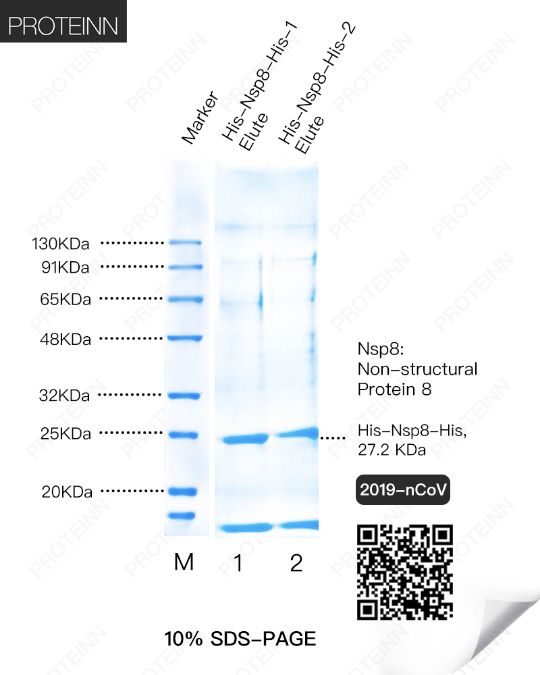
Nsp8: Non-Structural Protein 8
Function: Forms a hexadecamer with Nsp7 (8 subunits of each) that may participate in viral replication by acting as a primase. Alternatively, may synthesize substantially longer products than oligonucleotide primers.
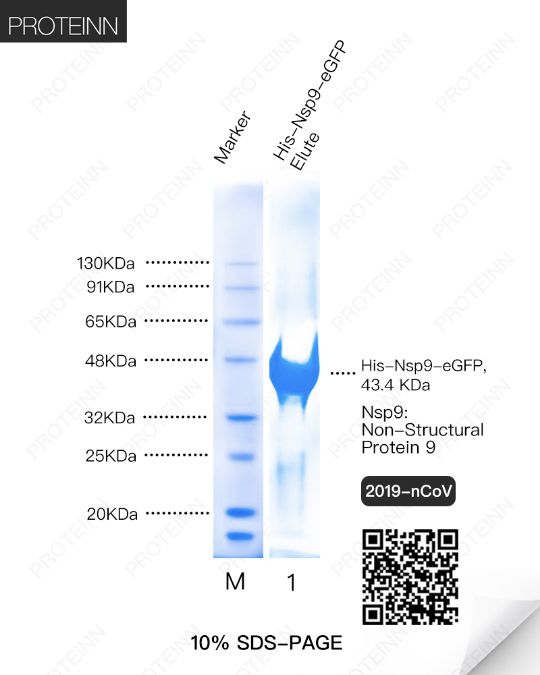
Nsp9: Non-Structural Protein 9
Function: May participate in viral replication by acting as a ssRNA-binding protein.
Nsp10: Non-Structural Protein 10
Function: Plays a pivotal role in viral transcription by stimulating both nsp14 3'-5' exoribonuclease and nsp16 2'-O-methyltransferase activities. Therefore plays an essential role in viral mRNAs cap methylation.
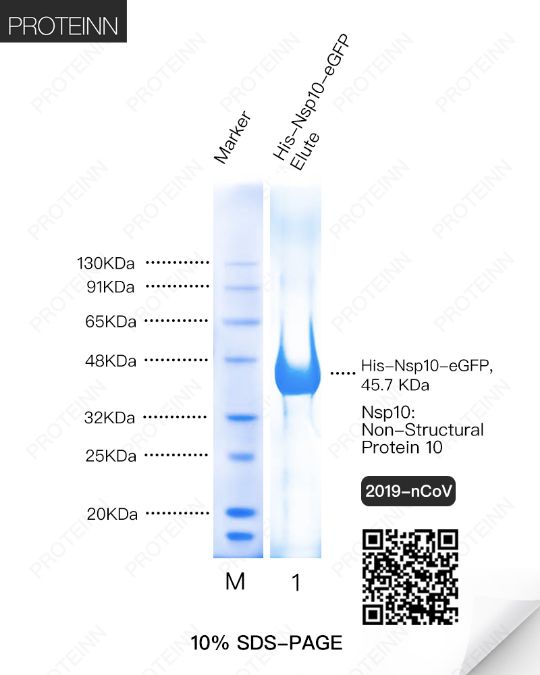

Table 1. 2019-nCoV Viral Protein List
The sequence lists were last updated Wednesday Feb 18 10:45 2020 EST, and are updated as additional sequences are released. The table content is available for download.
References.
1. Wuhan seafood market pneumonia virus isolate Wuhan-Hu-1, complete genome. NCBI Reference Sequence: NC_045512.2
https://www.ncbi.nlm.nih.gov/nuccore/179817425
2. Durai, P., Batool, M., Shah, M. et al. Middle East respiratory syndrome coronavirus: transmission, virology and therapeutic targeting to aid in outbreak control. 2015 Exp Mol Med 47, e181. PMC4558490
3. Experiments performed by NC Zhang, YL Zhao, XL Yang, Xue Y; Data Analysis by TT Hang, NC Zhang, XD Xu, ZN Huang, XQ Ju, M Guo, Kangma-Healthcode.com
4. Full Cell-Free Made Proteins of 2019-nCoV Information can be found on OurProteinFactory.com/2019-nCoV/protein